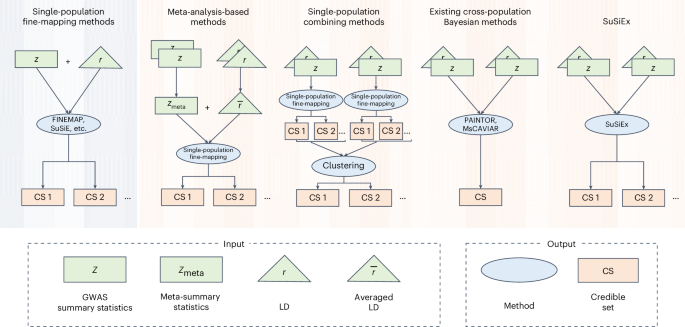
Fine-mapping across generations to stimulate the discovery of diverse causes underlying human processes and diseases – Nature Genetics
Huang, H. et al. Fine-mapping inflammatory bowel disease loci in a single-gene solution. Nature 547173-178 (2017).
Maller, JB and others. Bayesian optimization of association markers for 14 loci in 3 common diseases. Nat. Genet. 441294–1301 (2012).
Cortes, A. et al. Identification of multiple risk factors for ankylosing spondylitis by high-throughput genotyping of neural-associated loci. Nat. Genet. 45730–738 (2013).
Trubetskoy, V. et al. Mapping genomic loci involving genetics and synaptic biology in schizophrenia. Nature 604502–508 (2022).
Mahajan, A. et al. Genetic studies of multiple types of type 2 diabetes highlight the potential of different individuals for discovery and translation. Nat. Genet. 54560–572 (2022).
Kanai, M. et al. Meta-analysis fine-mapping is often mistaken for a single-model decision. Cell Genome. 2100210 (2022).
Kanai, M. et al. Data from maps of complex patterns across different populations. Printing before medRxiv https://doi.org/10.1101/2021.09.03.21262975 (2021).
LaPierre, N. et al. Identifying causal variables by fine-mapping across multiple subjects. PLoS Genet. 17e1009733 (2021).
Kichaev, G. & Pasaniuc, B. Leveraging data-annotation data in fine ethnic mapping studies. Am. J. Hum. Genet. 97260-271 (2015).
Wyss, AB and others. A multi-ethnic meta-analysis identifies a specific locus for lung function. Nat. Normal. 92976 (2018).
Gharahkhani, P. et al. Genome-wide meta-analysis identifies 127 open-angle glaucoma loci with similar effect across ancestry. Nat. Normal. 121258 (2021).
Robertson, CC and others. Mapping, trans-ancestral and genomic studies identify different causes, cells, genes and drug targets for type 1 diabetes. Nat. Genet. 53962–971 (2021).
Wang, G., Sarkar, A., Carbonetto, P. & Stephens, M. A new simple method for differential selection by regression, using fine genetic mapping. JR Stat. Soc. Statistical Series B. Method. 821273–1300 (2020).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of aging and aging PLoS Med. 12e1001779 (2015).
Feng, Y.-CA et al. Taiwan Biobank: a rich biomedical research site for Taiwanese citizens. Cell Genome. 2100197 (2022).
Mitchell, TJ & Beauchamp, JJ Bayesian variable selection using regression. J. Am. Stat. Assoc. 831023–1032 (1988).
George, EI & McCulloch, RE Bayesian variable selection methods. Stat. Sin. 7339–373 (1997).
Google Scholar
Su, Z., Marchini, J. & Donnelly, P. HAPGEN2: modeling multiple SNP diseases. Bioinformatics 272304–2305 (2011).
Auton, A. et al. Global reference for human genetic variation. Nature 52668-74 (2015).
Kichaev, G. et al. Integrating performance data prioritizes alternative causal models in fine-mapping statistical studies. PLoS Genet. 10e1004722 (2014).
Ulirsch, JC and others. Analysis of human hematopoiesis in single cell and differential solutions. Nat. Genet. 51683–693 (2019).
Weissbrod, O. et al. Systematic mapping and polygenic localization of complex heritability patterns. Nat. Genet. 521355–1363 (2020).
Ulirsch, JC Identification and Interpretation of Causal Genetic Variants Underlying Human Phenotypes. Doctoral thesis, Harvard University. Graduate School of Arts and Sciences (2022).
Chen, C.-Y. and al. Analyzes across Taiwan Biobank, Biobank Japan and UK Biobank identify hundreds of new loci for 36 quantitative traits. Cell Genome. 3100436 (2023).
Li, H. Toward a better understanding of cross-talk artifacts from high-resolution sampling. Bioinformatics 302843–2851 (2014).
Karczewski, KJ and others. The conversion rate was calculated from 141,456 people. Nature 581434–443 (2020).
Benner, C. and others. FINEMAP: an efficient alternative selection using a summary of data from genome-wide association studies. Bioinformatics 321493–1501 (2016).
Benner, C. and others. Prospects for fine-mapping genomic regions associated with traits using aggregate statistics from genome-wide association studies. Am. J. Hum. Genet. 101539–551 (2017).
Taliun, D. et al. Sequences of 53,831 different genes from the NHLBI TOPMed Program. Nature 590290–299 (2021).
Zhou, W. et al. Global Biobank Meta-analysis Initiative: enhancing gene discovery across human diseases. Cell Genome. 2100192 (2022).
Pruim, RJ and others. LocusZoom: a landscape view of genome-wide association analysis results. Bioinformatics 262336–2337 (2010).
Wang, QS et al. Using controlled learning to create an accurate knowledge map of cis-eQTL identifies 20,913 additional causal eQTLs. Nat. Normal. 123394 (2021).
Hou, Y. et al. Schizophrenia-associated rs4702 G down-regulation FURIN expression by miR-338-3p reduces BDNF production. Schizophr. Res. 199176–180 (2018).
Schrode, N. et al. Synergistic effects of common risk factors in schizophrenia. Nat. Genet. 511475–1485 (2019).
Manichaikul, A. et al. Dynamic correlation in genome-wide association studies. Bioinformatics 262867–2873 (2010).
Chang, CC and others. Second generation PLINK: meets the challenge of larger and richer databases. Gigascience 47 (2015).
Abraham, G., Qiu, Y. & Inouye, M. FlashPCA2: principal component analysis of Biobank-scale genotype datasets. Bioinformatics 332776-2778 (2017).
Han, B. & Eskin, E. Random-effects model for discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 88586–598 (2011).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17122 (2016).
Willer, CJ, Li, Y. & Abecasis, GR METAL: a rapid and efficient meta-analysis of genomewide association fragments. Bioinformatics 262190–2191 (2010).
Ge, T. & Yuan, K. getian107/SuSiEx: SuSiEx-v1.1.2 (v1.1.2). Zenodo https://doi.org/10.5281/zenodo.11211744 (2024).
#Finemapping #generations #stimulate #discovery #diverse #underlying #human #processes #diseases #Nature #Genetics